How to Read the Crystal Ball Accurately
Introduction
Definition of Foodome
In the present "omics" era, ane may believe based on literature and instrumental developments that a comprehensive motion-picture show of metabolites is known and tin can exist analyzed. However, upon closer inspection and despite immense technological developments, the major fraction still remains unknown. The question arises every bit to whether a forecast is possible and, if so, when a real comprehensive picture of food metabolites could be available.
Based on the definition of "foodomics" (i), nosotros recently specified "foodome" as being the "collection of all compounds present at a given time in an investigated nutrient sample and/or in a biological system interacting with the investigated nutrient" (two).
As a fraction of the foodome, food metabolome is the prepare of metabolites in nutrient and is supposed to exist accessible using land-of-the-art metabolomics platforms.
The food metabolome has been investigated in a targeted way since the by decades, just the last years have seen an increasing number of efforts to investigate the metabolome by non-targeted methods through applying electric current high-resolution platforms. Among these, eastward.yard., mass spectrometric detectors have already been compared lately (2).
Besides the continuous efforts to increase the mass exactness and resolving power of a mass spectrometer (MS), developments have been very successful in decreasing the limits of detection (LOD) down to the lower nanogram-per-kilogram level or beyond. The whole analytical pipeline will decide the sensitivity limitation of an analytical arroyo—from sample conditioning, extraction, and separation toward ionization source to ion manipulations and detections [in the case of liquid chromatography (LC)/MS]. Inside all targeted metabolomics platforms, the triple quadrupole (QQQ) mass detectors are now-a-days still ahead in sensitivity compared to fourth dimension-of-flight (TOF) detectors for the aforementioned food sample prepared identically.
Food metabolites are present in nutrient over a wide dynamic range of concentration (from attomolar to molar), and thus their possible detection depends on their individual amount in the considered nutrient, their possible selective isolation out of the matrix in chromatography, their selective ionization potential in the source (and suppression effects with the matrix), and the sensitivity of the mass detector in the mass range observed. Well-known metabolites thus might not be detected by not-targeted nutrient metabolomics although they are nowadays. For instance, the whole group of folate vitamers mostly is missing in a non-targeted study (3), merely in the validation of targeted folate methods, nosotros could hardly find whatsoever blank natural material that is gratuitous from folates (4). Moreover, in our studies on the metabolome of the Alternaria fungi, more than fifty% of the primary metabolites known in databases are still missing (v).
Simply a minority of instrumentally detected molecules are found in databases, not mentioning the isobars and isomers that frequently are not resolved in the analytical approaches (half-dozen).
These findings point to the being of unlike categories of "unknown" metabolites or dissimilar kinds of metabolic "dark matters" (seven).
Nosotros encounter currently a fast development of the analytical equipment (viii), and the increase in sensitivity and MS resolution appears to exist growing exponentially, which indicates the hypothesis of applicability of the Laws of Moore and Kurzweil in describing the general development of technology over time. In the early on 1960's, M.Eastward. Moore hypothesized that the number of transistors in an integrated circuit doubles every 2 years (9), and based thereon, in 1999 Ray Kurzweil proposed the Law of Accelerating Returns.
According to this police, the technological advancement will lead to "…a future catamenia during which the pace of technological change will be so rapid, its bear on and so deep, that human life will exist irreversibly transformed" (x). This menstruum corresponds to the "singularity" or, more precisely, the "technological singularity."
We hypothesize in this essay that all of these considerations mentioned in a higher place lead to the supposition that the laws of Moore and Kurzweil are too applicable to current metabolomics approaches and so to nutrient analysis. Therefore, the application of these laws may permit united states to predict roughly when all metabolites in foods will be (1) known, (two) detectable, and (3) identifiable. These predictions are the aims of the present viewpoint. It has to be highlighted that we limit our hypotheses to the development of mass spectrometry with its superior sensitivity and versatility, whereas other spectrometric methods are out of the scope of this study and certainly modulate the outcome in describing the chemic infinite (xi).
Underlying Data and Categories for Prediction
Predicting the Magnitude of the Metabolome
If nosotros want to predict the time when the whole nutrient metabolome volition be identified, we take to estimate outset the approximate number of metabolites that we expect to be identified. Considered herein as "metabolites" are but those direct biological pocket-sized molecule metabolites of living edible systems, i.e., establish, animal, and microorganism sources—not considered are compounds of chemical abiotic origin or from chemical transformations/recombinations (eastward.g., hydrolysis, thermolysis, Maillard reaction). For the estimation of these compounds, a look into contemporary metabolite databases is the starting time step. Several compound databases from different organizations or consortia have been published with different foci in the terminal decade. A brusk overview about the number of compounds, focus, and publisher or curator is given in Table 1.

Table one. Examples of important metabolite databases.
The number of compounds included in these databases ranges from a few thousands (GMD) to almost 100 1000000 (PubChem)—the sources ranging from principal metabolites in certain species like humans to all man-fabricated chemical species, respectively. In principle, all of these compounds may be occurring in foods, merely the bulk of them, being xenobiotica, have very low probability of advent. For an interpretation of the most likely metabolites generally to be expected in foods, information technology is straightforward to exclude, in a first approximation, all xenobiotica. Moreover, from recent studies on non-enzymatic browning during thermal processing of foods, besides termed Maillard reaction, information technology became obvious that this reaction network results to several tens of thousands of boosted metabolites that accept started from just a few reactive compounds (eleven–14). Apart from these, many further reactions of food metabolites similar thermolysis, hydrolysis, or the plethora of lipid peroxidation reactions tin be causeless to increase the number of metabolites tremendously. In lodge to proceed the numbers manageable, it appears straightforward to merely focus on the endogenous compounds of the principal and secondary metabolism of plants, animals, and microorganisms occurring in non-processed foods. This as well includes their so-called stage ane and 2 metabolites. Similarly like not considering metabolites arising from processing such equally from Maillard reaction, PubChem, ChemSpider, and Metlin should non be included as they mainly contain xenobiotica. A much improve estimation is based on the ~twenty,000 compounds in KEGG or the currently updated homo metabolome database (xv) containing 114,000 compounds. For the institute metabolome, more 200,000 compounds have been estimated (sixteen). Regarding lipids, the most comprehensive database LIPID MAPS lists effectually 44,000 compounds with in-silico predictions running into the hundreds of thousands (17). Some entries of these databases are overlapping, only the extent of overlap is hard to estimate (xviii). In lodge to follow a rather worst-instance scenario, bold that there are possibly additional 100,000 notwithstanding hitherto unknown metabolites, a rough estimation for our most conservative prediction may exist 500,000 as the number of all relevant food metabolites. From this rough deduction, the magnitude of uncertainty is obvious and the true number may range from one-half of this figure to its double, i.e., some hundreds of thousands of additional possible and believable metabolites not covered still.
The next stride and assumption for our prediction volition be the course of fourth dimension in which these compounds will be discovered. Every bit outlined by Kurzweil (10), many developments, particularly in calculating and data treatment, follow an exponential evolution. When having a brusque look at the developments of the databases mentioned above, this time evolution has been reported for HMDB in 2018 and is visualized in Figure 1A. It is evident that, between the years 2007 (HMDB 1.0) and 2018 (HMDB 4.0), the number of compounds increased in this predicted exponential way (15) and tin be farther projected appropriately into the future when assuming the Law of Accelerating Returns (10). A farther information from HMDB can exist ended, i.e., (i) the number of predicted compounds, (ii) those that have been detected, and (3) those that accept been quantified. This indicates several categories of "unknowns" or "dark matters," which will exist further hypothesized and outlined below.
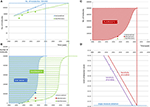
Effigy 1. Development and prediction of predicted, detected, and identified metabolites over fourth dimension. (A,B) The number of predicted and identified compounds in the HMDB versions 1.0 (2007) and 4.0 (2018) (15) increased in the given fourth dimension range in a predicted exponential style and tin exist further projected accordingly into the futurity to reach the number of 500,000 metabolites comprising the relevant metabolites in foods, excluding xenobiotica and process-generated compounds. The fractions of not-predicted and not-identified metabolites are termed dark matter I and night matter Iii, respectively. (C,D) The evolution of point sensitivity over time in gimmicky LC-QQQ MS instrument in (D) is translated into a limit of detection for the given injected amount of molecules, hither for the reference compound reserpine. Nether the assumption that the limit of detecting reserpine refers to a constant threshold of signal intensity, the limit of detection has been decreasing over fourth dimension. The evolution follows an exponential behavior over the terminal 8 years and can exist predicted to follow this path in the future until unmarried molecule detection is reached. As the sensitivity of non-targeted LC-QTOF MS can be expected to exist i order of magnitude lower than that of the LC-QQQ, single molecule detection of the sometime volition exist reached later. The respective yr projected to the current state of detected molecules in HMDB and the estimation of the further evolution to detect the expected number of 500,000 metabolites result in "dark matter 2," which is equivalent to the non-detected metabolites over time.
Predicting the Performance of Belittling Equipment
In order to find, identify, and quantify the predicted number of metabolites, the adjacent supposition for our prediction is the forecast on how the performance of the analytical equipment in spectroscopy, spectrometry, and separation sciences, as the analytical pillars, will develop (11). In this respect, we commencement have to differentiate between targeted and not-targeted metabolomics platforms as outlined above. Second, we have to compile the relevant key performance indicators (KPIs) of the instrumentation such as sensitivity of detection, chromatographic resolution, mass accuracy or MS resolution, and their development over time. For both targeted and non-targeted metabolomics, the primary KPI is the sensitivity of detection, and compiling these data for all analytical approaches would exist challenging. As each analytical technology has its own criteria to document sensitivity, for this review, the evolution of sensitivity already published for one analytical platform (2) has been connected.
For instance, development in the sensitivity of mass spectrometric equipment is referenced for the chemical compound reserpine every bit the signal intensity for a given amount injected into the MS. The ionization efficiency for whatever other chemic tin can be dissimilar by upward to many multitudes and makes information technology difficult to generalize, but a farther assumption to extend our hypothesis volition exist that the limit of detecting reserpine refers to a abiding threshold of signal intensity, which means that the limit of detection is decreasing over time as revealed in Figure 1D.
It is over again obvious that the development follows an exponential behavior over the last 8 years and can exist predicted to follow this path in the future when assuming the Police of Accelerating Returns fix past Kurzweil. Considering not-targeted LC-QTOF instrumentation, a certain gap in sensitivity is obvious when compared to LC-QQQ, simply the equivalent development of increasing sensitivity over time tin be assumed. Again it has to be stressed that, within manufacturers' equipments, we will observe different performances in sensitivity, only it tin be assumed that all of them fall in the same order of magnitude; otherwise, they would not take been competitive in the market up to present.
The side by side assumption refers to the target sensitivity that the equipment would have to attain in our prediction. In this respect, a comprehensive detection is required, i.e., a metabolite should be detectable if only 1 molecule is present in the respective amount of food. This is equivalent to the requirement of single molecule detection. It is articulate that the currently detectable few femtograms of reserpine still equals effectually a few million molecules, simply this number tin be expected to exponentially subtract according to the trend of the terminal years (see below) and the assumed Police of Accelerating Returns.
A further KPI to consider would be mass spectrometric resolution, and the exponential increase for TOF instruments has already been reported by Bristow (19).
When comparing the existing lack in sensitivity and resolution of contemporary instruments, the requirement to bridge the sensitivity gap of more six orders of magnitude toward single molecule detection on the footing of reserpine appears more challenging than the necessity to resolve all metabolites. The resolution currently achieved by the Orbitrap-blazon of instruments is in the several hundreds of thousands, and 21 Tesla FTICR-MS instruments may already reach up to 50,000,000 and this resolution appears higher than currently needed for nutrient metabolites. Moreover, analytical resolution can too be increased in systems chemical analytics with hyphenations of mass spectrometry with separation sciences or spectroscopy equipments in various dimensionality (i.e., LC-HRMS-NMR or GCxGC-HRMS) (eleven, 20, 21). Sufficient resolution is already or will soon be achieved inside the time range until unmarried molecule detection will be available; unmarried molecule imaging is already reached with techniques such as diminutive force microscopy.
Predicting Analytical Singularity
For this prediction, assuming several hypotheses, the time development of the developments in detecting, identifying, and quantifying the metabolome has to be followed up in the futurity. The target number of metabolites would be hypothetically set at 500,000 in the present study to enable a first evaluation, and when projecting the development of the predicted metabolites in the HMDB, the progress displayed in Figure 1B can be assumed with its exponential projection asymptotically reaching the number of 500,000 in ~2025. This ways that, based on such a model, there should no longer be whatever unknown relevant metabolites in 2025, which appears rather unrealistic when because the electric current low percentages of the assigned metabolites in metabolic studies. Until and then, these currently unknown compounds may be assigned to the first category of "dark matter," i.e., "unknowns" or "nighttime matter I" (Effigy 1B).
The next prediction refers to the time when all of these metabolites will be detectable past analytical equipment as resolved features, and this requires the project of the not-targeted metabolomics to single molecule detection. This is outlined in Figure 1D, which projects the exponential evolution of the current LOD of vi × 10−eighteen mol (Table S1) to the LOD beingness one molecule, i.e., one.66 × 10−24 mol. Assuming this model, single molecule detection on routine MS equipment should exist feasible by around 2032. It has to be mentioned that unmarried molecule detection is already bachelor in unmarried particle mass spectrometry (22) or in mod atomic strength microscopy (23) with which even the aliphatic or aromatic rings can be visualized. In conclusion, the second category of dark matter would refer to "not-detected" or "dark thing II" and is indicated in Effigy 1C. When comparing "night matter I" with "nighttime matter 2," it has to be kept in listen that the detected molecules may be either unknown or known, i.e., not being present in databases and belonging to "dark affair I" or being nowadays in databases and belonging to not-"dark thing I," respectively. Therefore, "nighttime matter I" is not a consummate subset of "night affair Two."
Several weaknesses of this prediction have to be admitted: first, the possibility of single molecule detection does not necessarily imply that all unmarried molecules in a circuitous food extract will be detectable due to ionization selectivity in mass spectrometry, high dynamic ranges in abundances, presence of enantiomers, etc. As we have to consider ionization suppression for virtually all ion sources, the current process to circumvent this would be dilution, but this would require an even higher sensitivity. This ways that the time point to achieve single molecule detection would be even later, if e'er. Apart from general sensitivity considerations, we yet have to go along in mind that contemporary MS instrumentation may hardly discover the classes of compounds that are hardly ionizable, and uncovering these will require novel kinds of ion sources. Multiple ionization sources may be useful to comprehend a broader chemical range and to observe more compound classes. We showed, e.k., in a study with FTICR-MS of complex organic mixtures involving ESI, APPI, and APCI in both positive and negative ionization modes that electrospray ionization in positive mode covers simply xxx% of the total appreciable compositional space involving all modalities (half dozen). Moreover, the need to differentiate between all stereoisomers points to the demand of novel methods or chromatographic systems for enantioseparation.
Predicting the Identification of the Whole Nutrient Metabolome
Moreover, one molecule appearing as a resolved analytical signal in the analysis does not necessarily hateful that the chemical compound is already identified in structure. In dissimilarity to NMR, MS will only provide the elemental formula and, therefore, let the molecule to be assigned but tentatively to an unequivocal chemical structure. Moreover, in that location volition still be ambivalence about the identity of the compound fifty-fifty when MS/MS fragmentation, for instance, is practical and gives further evidence most the putative structure. This limitation directly leads to the definition of a 3rd level of dubiety, i.e., the "not-identified" or "night matter III." When comparison "nighttime matter 3" with "dark thing I" and "dark affair II," it is articulate that identified molecules are necessarily known and detected, i.e., members of "dark matter III" are either components of "night matter I" or "night matter 2" or of both.
For unraveling "dark matter III," the prediction is fifty-fifty more than hard and more discipline to high fluctuations as the straight projection would crave data on developments of either all metabolites' extraction, clean-upward, and spectroscopic structural assignment by, i.e., NMR or chemical synthesis (if possible in a reasonable time) and commercial availability to that date. However, a rather coarse prediction is possible from the already mentioned HMDB database and here from the number of identified and quantified metabolites developing over the last decade (green curve in Figure 1A). These numbers are much smaller than those of the other categories, and it will take much longer to completely unravel dark matter Three. If the projection is assumed to be a combination of the current exponential evolution and to run in parallel with the projection of dark matter I and 2, the time betoken when all food metabolites will be assignable in food samples could be expected in this model by around 2041 (Effigy 1B).
Discussion
The upmost weakness of the prediction presented hither is its ambiguity and requires an intense uncertainty assessment and permit united states play with numbers. This starts already with the uncertainties referring to the number of metabolites upward to the evolution of the unequivocally identified metabolites. The assumed dubiousness of l% of the existing number of metabolites would already atomic number 82 to a time range from 2022 to 2028, respectively, for unraveling dark matter I. The speculated uncertain number of 500,000 different metabolites is also relatively conservative, and assuming a number of one 1000000 would already lead to 2028 for revealing nighttime matter I and 2060 for nighttime affair II. This is already without considering the plethora of all conceivable metabolites based on chemical science rules that can reach extremely high numbers (xi). For instance, a molecule consisting only of carbon, hydrogen, and oxygen with the formula CnH2nOn at a molecular mass of 500 would have theoretically around x16 calculated possible isomers—and with the formula CnorthwardHnorthOnorth an even college number of x20 isomers, respectively (xi). The magnitude of all possible reactions, e.g., during processing or storage (abiotic degradations such every bit reductions/oxydations, condensation reactions, and polymerizations), is not even counted in this very conservative hypothetical assay. The sensitivity differences betwixt the aforementioned type of musical instrument from dissimilar manufacturers may cover one order of magnitude, and the necessity to further dilute the extracts to overcome ionization suppression has an impact on our calculations and thus may expand the time range for uncovering dark thing Ii (i.e., single molecule detection) from 2029 to 2038. For dark thing Three, the time prediction may range from a projection running in parallel with the best-case scenario for dark matter II, i.eastward., around 2035, to a flat exponential projection of quantified metabolites in HMDB from the database'southward last update approximating the number of 500,000 not revealed before the end of the twenty-get-go century. For this development, the law of Kurzweil obviously does not apply inside the observed fourth dimension menstruum. This exemplifies the well-known proverb attributed to Niels Bohr who said that "prediction is very hard, particularly nearly the time to come" (24) and thus sets the real limitation of our wizzard's crystal ball's reading.
From all of the considerations mentioned above, it seems clear that, in particular, 3 bottlenecks take to be circumvented earlier the whole nutrient metabolome can be unraveled: (i) coverage and curation of databases, (ii) sensitivity and resolution of analytical equipment, and (three) unequivocal metabolite identification.
These issues have to be addressed past the food analytical customs, including researchers and developers at academic institutions and instrument manufacturers. A current target for the latter would be the evolution of ultrasensitive and ultrahigh resolution equipment (in mass spectrometry and multidimensional chromatography). We hope that the law of Kurzweil will be applicable to all of these methodologies.
Further aspects in foodomics involving the quantitation of all of these metabolites have not been mentioned above. This hopefully will be achievable sometime after "all" metabolites have been identified as, with the availability of the needed reference compounds or with the knowledge of unequivocal belittling properties, targeted methods volition exist bachelor within the next decades.
Another aspect which has been often discussed with respect to analytical development is the miniaturization of the equipment with reduced need in sample amount and the consistent reduction of analysis time. These properties tin be expected to also show a similar exponential evolution and would lead to fast, portable, and selective sensors, the evolution of which is likewise currently taking place. However, this prediction is out of the telescopic of this perspective.
At present, all three dark matters exist meantime and are recognizable for any analytical chemist working in metabolomics, e.yard., foodomics. In the further years, the follow-up of the development in noesis predicted for the three dark matters mentioned herein, with the predictions presented hither, will indicate the validity of this perspective.
When comparison these predictions with those of the singularity in artificial intelligence (AI) and human intelligence predicted for 2045, it appears likely that by and then we will take unraveled at to the lowest degree dark matter I and dark thing II, and the latter singularity may speed upward the unraveling of the remaining dark matter 3. The help of AI may even have some unpredictable outcome on the speed of discoveries and unraveling of all dark affair. Besides, after singularity has been reached, the world every bit we know it now may completely have changed in many other different respects every bit well.
Data Availability Argument
All datasets generated for this study are included in the article/Supplementary Fabric.
Writer Contributions
All authors listed have made a substantial, straight and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the enquiry was conducted in the absence of any commercial or fiscal relationships that could be construed as a potential conflict of involvement.
Supplementary Material
The Supplementary Cloth for this commodity tin can be plant online at: https://www.frontiersin.org/manufactures/ten.3389/fnut.2020.00009/full#supplementary-fabric
References
ii. Rychlik G, Kanawati B, Schmitt-Kopplin P. Foodomics every bit a promising tool to investigate the mycobolome. Trends Anal Chem. (2017) 96:22–xxx. doi: 10.1016/j.trac.2017.05.006
CrossRef Full Text | Google Scholar
3. Rychlik M, Kanawati B, Roullier-Gall C, Hemmler D, Liu Y, Alexandre H, et al. Foodomics assessed by fourier transform mass spectrometry. In: Kanawati B, Schmitt-Kopplin P, editors. Fundamentals and Applications of Fourier Transform Mass Spectrometry. Amsterdam: Elsevier (2019). p.651–77.
Google Scholar
4. Mönch Southward, Rychlik Thousand. Improved folate extraction and tracing deconjugation efficiency by dual label isotope dilution assays in foods. J Agric Food Chem. (2012) 60:1363–72. doi: ten.1021/jf203670g
PubMed Abstract | CrossRef Total Text | Google Scholar
5. Gotthardt Thou, Kanawati B, Schmitt-Kopplin P, Rychlik M. Comprehensive analysis of the metabolome of alternaria isolates by FT-ICR-MS and LC-MS/MS. Mol Nutr Food Res. (2019) 64:e1900558. doi: 10.1002/mnfr.201900558
CrossRef Full Text
half dozen. Hertkorn N, Frommberger Grand, Witt M, Koch BP, Schmitt-Kopplin P, Perdue EM. Natural organic thing and the result horizon of mass spectrometry. Anal Chem. (2008) 80:8908–19. doi: x.1021/ac800464g
PubMed Abstruse | CrossRef Total Text | Google Scholar
8. Rychlik M, Gotthardt M, Kietz R, Maurer A, Gunkel K, Asam S, et al. Alternaria-toxine treten in Erscheinung. Nachr Chem. (2018) 66:877–80. doi: 10.1002/nadc.20184071730
CrossRef Full Text | Google Scholar
10. Kurzweil R. The Singularity is Near. New York, NY: Viking Press (2005).
Google Scholar
eleven. Schmitt-Kopplin P, Hemmler D, Moritz F, Gougeon RD, Lucio Grand, Meringer Grand, et al. Systems chemic analytics: introduction to the challenges of chemical complication analysis. Faraday Discuss. (2019) 218:9–28. doi: 10.1039/C9FD00078J
PubMed Abstract | CrossRef Full Text | Google Scholar
12. Hemmler D, Gonsior M, Powers LC, Marshall JW, Rychlik M, Taylor AJ, et al. Faux sunlight selectively modifies maillard reaction products in a wide assortment of chemical reactions. Chem A Eur J. (2019) 25:13208–17. doi: 10.1002/chem.201902804
PubMed Abstract | CrossRef Full Text | Google Scholar
13. Hemmler D, Roullier-Gall C, Marshall JW, Rychlik M, Taylor AJ, Schmitt-Kopplin P. Insights into the chemistry of non-enzymatic browning reactions in different ribose-amino acrid model systems. Sci Rep. (2018) 8:16879. doi: 10.1038/s41598-018-34335-5
PubMed Abstract | CrossRef Full Text | Google Scholar
14. Hemmler D, Roullier-Gall C, Marshall JW, Rychlik M, Taylor AJ, Schmitt-Kopplin P. Evolution of complex maillard chemical reactions, resolved in time. Sci Rep. (2017) 7:3227. doi: 10.1038/s41598-017-03691-z
PubMed Abstruse | CrossRef Total Text | Google Scholar
15. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang Grand, Vázquez-Fresno R. HMDB 4.0: the homo metabolome database for 2018. Nucleic Acids Res. (2018) 46:D608–17. doi: x.1093/nar/gkx1089
PubMed Abstract | CrossRef Full Text | Google Scholar
eighteen. Vinaixa Yard, Schymanski EL, Neumann Southward, Navarro Grand, Salek RM, Yanes O. Mass spectral databases for LC/MS- and GC/MS-based metabolomics: state of the field and future prospects. Tr Anal Chem. (2016) 78:23–35. doi: 10.1016/j.trac.2015.09.005
CrossRef Total Text | Google Scholar
20. Hertkorn Northward, Ruecker C, Meringer K, Gugisch R, Frommberger M, Perdue EM. Loftier-precision frequency measurements: indispensable tools at the cadre of molecular-level assay of complex systems. Anal Bioanal Chem. (2007) 389:1311–27. doi: x.1007/s00216-007-1577-4
PubMed Abstruse | CrossRef Full Text | Google Scholar
21. Forcisi S, Moritz F, Kanawati B, Tziotis D, Lehmann R, Schmitt-Kopplin P. Liquid chromatography-mass spectrometry in metabolomics inquiry: mass analyzers in ultra high pressure liquid chromatography coupling. J Chromatograp A. (2013) 1292:51–65. doi: x.1016/j.blush.2013.04.017
PubMed Abstract | CrossRef Full Text | Google Scholar
23. Iwata M, Yamazaki S, Mutombo P, Hapala P, Ondracek M, Jelinek P, et al. Chemic structure imaging of a single molecule by atomic force microscopy at room temperature. Nat Commun. (2015) six:7766. doi: 10.1038/ncomms8766
PubMed Abstract | CrossRef Full Text | Google Scholar
24. Restriction M. The Science of Science Fiction. New York, NY: Simon and Schuster (2018).
Google Scholar
Source: https://www.frontiersin.org/articles/516095
0 Response to "How to Read the Crystal Ball Accurately"
Publicar un comentario